Kneading Enzyme Makes Ammonia Levels Rise
Electron-delivering protein manipulates natural catalyst, changing ideas about fertilizer production
(June 2015)
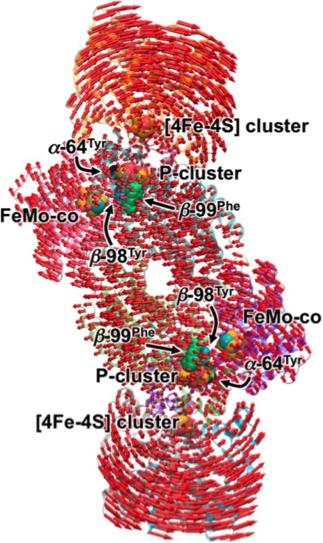
The rocking motion of the proteins ([4Fe-4S] cluster) is shown on the enzyme, with FeMo-co representing the active site. The length of the arrows is proportional to the displacement of the amino acid residues. Enlarge Image
Results: In industry, synthesizing ammonia for fertilizers uses massive amounts of hydrogen, typically generated from fossil fuels, but in nature, the nitrogenase enzyme produces ammonia without added hydrogen. In studying the enzyme, scientists came up against a protein, called the Fe protein. This little protein delivers electrons to the larger nitrogenase MoFe enzyme. The smaller protein's actions limit the enzyme's speed. Recently, scientists at Pacific Northwest National Laboratory and three universities found that the smaller protein and larger enzyme roll across each other, likely pushing at the MoFe surface to deliver electrons.
Why It Matters: Producing ammonia for the world's crop fertilizers consumes 1 to 2 percent of all the energy produced by humankind. Part of that energy is used to generate hydrogen gas, which is combined with nitrogen gas in the Haber-Bosch process. This research sheds new light on how an enzyme catalyzes a reaction without added hydrogen; instead, the enzyme uses protons and electrons.
Methods: The little Fe protein that delivers the electrons comes under scrutiny, as it is the dissociation of this protein from the MoFe protein or enzyme that is the slowest step in the process. And, the dissociation step must happen 8 times for each molecule of ammonia that is produced. Expediting or removing this rate-limiting step would provide fundamental insights into the nature of the enzyme, insights that could be applied to synthetic catalysts.
Substitution of three amino acids buried deep in the MoFe protein allows electrons to be delivered from small molecule electron carriers to support the transformation, or reduction, of several substrates, including azide to ammonia, hydrazine to ammonia and protons to molecular hydrogen, or H2.
A computational analysis indicated a mechanical coupling between the motions of the Fe protein and the region of the MoFe protein with these three amino acids, which suggests a possible mechanism for how Fe protein might communicate subtle changes deep within the MoFe protein that profoundly affect intramolecular electron transfer and substrate reduction.
The team found that the Fe protein does not merely dock with the enzyme to deliver an electron, but rather the protein massages the MoFe protein, rolling along the surface. This mechanical action directs the enzyme as well. The protein massage modulates the electron's journey through the catalyst to the active site.
The protein's movement as well as that of the enzyme's pushes the "stepping stones" used by the electrons to travel through the enzyme closer together. The result: faster ammonia production.
The kneading is an out-of-phase correlation, meaning the enzyme doesn't respond proportionately to the protein's original push. "This is a very unexpected and far from trivial result," said Dr. Simone Raugei, a PNNL scientist on the study.
This work was possible because of the collaborative interdisciplinary team assembled for both the experiments and the computational analysis. The team brought together experts in biology and proteins to characterize the enzymes along with experts in computational analysis, biophysics, and catalysis. The team was led by Dr. Lance Seefeldt, who has a joint appointment with PNNL and Utah State University. The research complements the physical biosciences work done at the national lab on the different aspects of the enzyme.
What's Next? Now, the researchers are delving into the details of the protein's influence on electron transfer providing information to delineate -- at a molecular level -- the mechanisms of electron delivery in a metal-based enzyme that catalyzes one of the most energy-demanding reactions. The work will inform electron and proton research at the Center for Molecular Electrocatalysis as well as protein interactions involved in physical bioscience research.
Acknowledgments
Sponsors: U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences (LCS, DRD) supported all of the protein work, while the Chemical Sciences, Geosciences, and Biosciences Division (SR) supported the computational work. The National Science Foundation (JWP and LCS) supported the X-ray crystallography.
User Facility: Stanford Synchrotron Radiation Lightsource
Research Areas: Biological Sciences, Chemical Sciences
Research Team: Karamatullah Danyal, Andrew Rasmussen, Boyd S. Inglet, Sudipta Shaw, Simon Duval, and Lance C. Seefeldt, Utah State University; Stephen M. Keable, Oleg A. Zadvornyy and John W. Peters, Montana State University; Dennis Dean, Virginia Tech University; Simone Raugei, PNNL
Reference: Danyal K, AJ Rasmussen, SM Keable, BS Inglet, S Shaw, OA Zadvornyy, S Duval, DR Dean, S Raugei, JW Peters, and LC Seefeldt. 2015. "Fe Protein-Independent Substrate Reduction by Nitrogenase MoFe Protein Variants." Biochemistry 54(15):2456-2462. DOI: 10.1021/acs.biochem.5b00140
